Microfluidic droplet systems allow the manipulation of small volumes of liquids with two immiscible phases, such as water and oil. The result is a small reactor in which a chemical reaction or biological process can be carried out and observed over time. The microdroplets can be mixed, sorted, incubated, and analyzed. These operations can be performed in specially designed microfluidic systems, creating a small lab-on-a-chip device. The main goal of our research is to observe the behavior of clinically relevant bacterial strains, particularly how they respond to antibiotics. Optics and laser technology combined with microfluidic systems allow us to conduct experiments much faster.
Antimicrobial resistance (AMR) is one of the world’s most pressing health threats. It occurs when bacteria, viruses, fungi, and parasites transform over time and no longer respond to drugs. As a result, antibiotics or other antimicrobial drugs become ineffective and fail to treat diseases. The World Health Organization (WHO) has identified AMR as one of the top 10 public health threats worldwide.
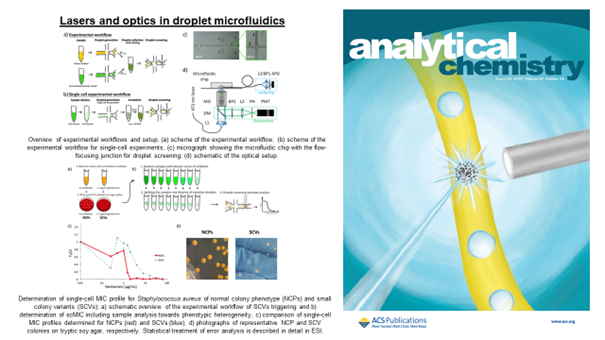
Monitoring the behavior of bacteria, i.e., their growth, is complex and time-consuming, especially when we have to keep track of thousands or millions of repeat experiments. Optical methods combined with microfluidics allow us to solve this problem. We can move droplets in front of a laser beam and analyze the light scattered on bacterial cells using specially designed chips. The intensity of the scattered light is related to the concentration of bacteria in the droplets, and we can track it over time. We can monitor over 1,000 droplets per second and analyze them with dedicated software. In addition, we can make the system more compact and easier to use by using fiber optics; we proposed a system in which a specially selected optical fiber is used to collect the light scattered on the bacteria [1].
Still, severe non-healing infections are often caused by multiple pathogens or genetic variants of the same pathogen exhibiting different levels of antibiotic resistance. For example, polymicrobial diabetic foot infections double the risk of amputation compared to monomicrobial infections. Although these infections lead to increased morbidity and mortality, standard antimicrobial susceptibility methods are designed for homogenous samples and are impaired in quantifying heteroresistance. We propose a droplet-based label-free method for quantifying the antibiotic response of the entire population at the single-cell level. We used Pseudomonas aeruginosa and Staphylococcus aureus samples to confirm that the shape of the profile informs about the coexistence of diverse bacterial subpopulations, their sizes, and antibiotic heteroresistance. These profiles could therefore indicate the outcome of antibiotic treatment in terms of the size of remaining subpopulations [2].
Author: Jakub Bogusławski, PhD
Team:
Jakub Bogusławski, PhD jboguslawski@ichf.edu.pl
Kamil Liżewski, PhD klizewski@ichf.edu.pl
Prof. Maciej Wojtkowski mwojtkowski@ichf.edu.pl
Publications:
- Natalia Pacocha, Jakub Bogusławski, Michał Horka, Karol Makuch, Kamil Liżewski, Maciej Wojtkowski, Piotr Garstecki, „High-Throughput Monitoring of Bacterial Cell Density in Nanoliter Droplets: Label-Free Detection of Unmodified Gram-Positive and Gram-Negative Bacteria,” Analytical Chemistry (2020).
- Natalia Pacocha, Marta Zapotoczna, Karol Makuch, Jakub Bogusławski, Piotr Garstecki, “You will know by its tail: a method for quantification of heterogeneity of bacterial populations using single-cell MIC profiling,” Lab on a Chip 22, 4317-4326 (2022).

